Abstract
Molecular analysis for studying the spatial configuration and properties of proteins has applications in modern biotechnology. In the present work, the enzyme pectate lyase (Pel1), obtained in the early stages using the transformation of recombinant plasmids into a bacterial culture of E. coli BL21(DE3), is studied in this way.
Background
Modern molecular biology has a sufficient variety of instrumental methods to thoroughly analyze the configurational properties of protein molecules and thereby predict their properties. The promise of such bioinstrumental analysis is extremely high since understanding the properties of molecules allows them to be manipulated for multiple applications, including biotechnology, pharmacology, medicine, and other dependent industries. Among advanced methods, X-ray crystallography stands out in particular, which allows the determination of the atomic and molecular structure of crystals by forming a diffraction pattern of rays deflected by the spatial shape of the protein crystal (Kermani, 2021). The comparative simplicity of the method, combined with the ability to study a diverse variety of proteins, has led to a scarcity of scientific materials with the potential to be used for crystallography emphasized in the literature. As a result, it creates a need for more in-depth studies to discover new protein molecules and to synthesize recombinant proteins that may also be of interest to study. Prominent among these is pectate lyase (Pel1), an extracellular enzyme that catalyzes the cleavage reactions of plant soft tissues (Wu et al., 2020). The application value of Pel1 is high, as this substance can be used by agricultural biotechnology to break down residual forms of starch in the alcohol industry and for food security needs. Production of a Pel1 protein crystal from a recombinant plasmid and subsequent analysis by X-ray crystallography are the objectives of this practical report.
Results
Spatial Structure Study
The central goal of the present study was the analytical study of the pectate lyase (Pel1) molecule obtained in the previous stages of the study. Previously, the substance was synthesized, assembled, and purified from Escherichia coli cells used as a vector agent for the transformation of plasmid pET-28a carrying the recombinant gene of this enzyme. The pure protein was obtained by lysing the incubated bacterial culture and then purifying by affinity chromatography: this allowed the separation of all protein molecules of the cells based on their interaction with transition metal ions. Crystallization screening of the obtained molecules was performed at a wavelength of 280 nm since, theoretically, this emission determines the maximum absorption of ultraviolet light (Quantifying protein, n.d.). This allowed the concentration of the enzyme to be calculated indirectly by Beer-Lambert law, after which the collected protein was crystallized. Figure 1 below shows the three-dimensional structure of this protein obtained by optical microphotography.
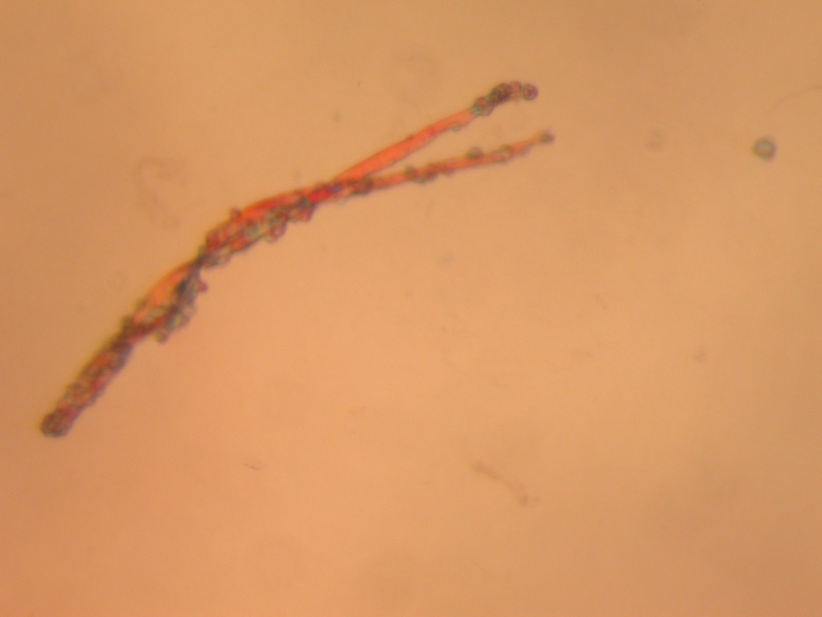
Molecular Weight Determination
In addition, one of the objectives of the current study was to measure the purity of the resulting protein crystal using the standard denaturing polyacrylamide gel electrophoresis method. This is a classic method of analytical biotechnology that allows the separation of proteins based on their electrophoretic mobility. However, the separation of proteins is not the final goal because due to the feature of the method, researchers are able to determine the relative molecular weights of the separated molecules and thus indirectly judge their qualitative composition. Figure 2 shows a molecular weight scale with numerical labels corresponding to the atomic mass units of the crystal. This scale was used to correlate the actual results with the reference labels, resulting in standard levels transferred to the real gel electrophoresis pattern for Pel1. As seen in Figure 3, the protein analyzed gave a signal at 40 kDa, indicating specifically this molecular weight inherent in Pel1.
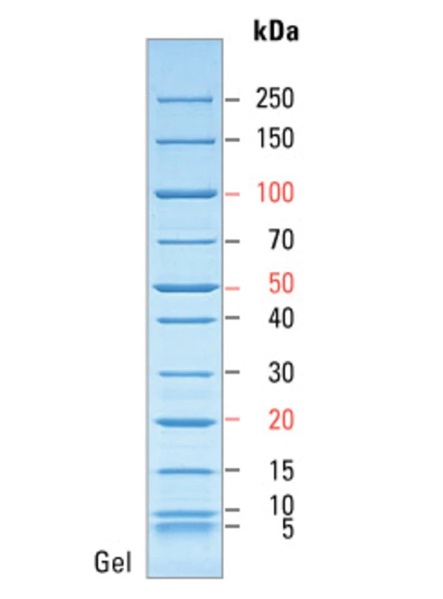
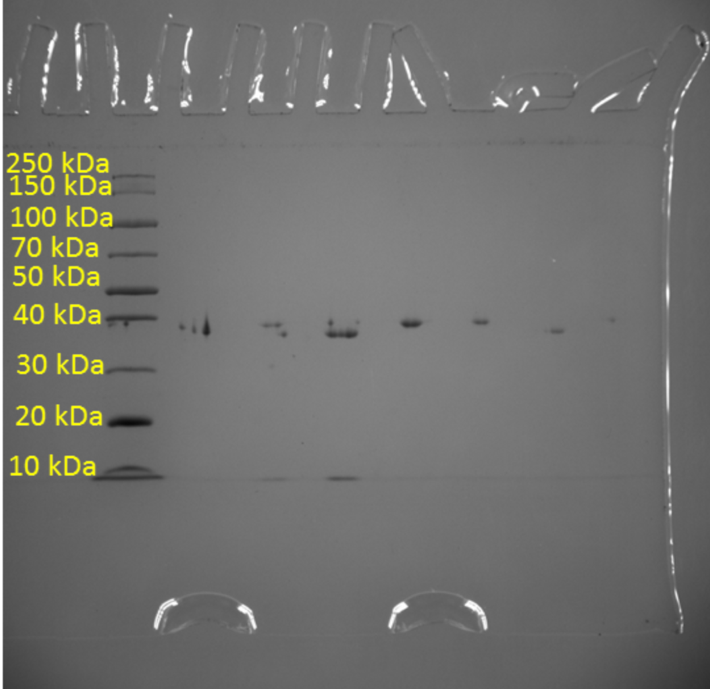
Discussion
The results obtained shed light on the morphological and weight properties of the isolated protein enzyme. In particular, the structure, shape, and approximate size of the Pel1 crystal were studied. Among the analytical results, the molecular weight of this compound was found to be 40 kDa. Reference to digital databases makes it clear that the results found are very close to the standards. In particular, UNIPROT reports a molecular weight for Pectate lyase Pel1 isolated from Ambrosia artemisiifolia of 43.665 Da (UniProt, 2021). Careful consideration of Fig. 3 makes it clear that the real weight of this molecule is extremely close to 40 kDa but does not reach this mark, which distances the obtained value from the reference standard. The probable reason for this phenomenon is the non-absolute purity of the protein and the presence of additive impurities that are firmly bound to the protein and reduce the total molecular weight.
Despite minor differences, the goal stated at the beginning was fully achieved as the molecular properties of the protein crystal were studied. The information gathered not only reflects the author’s competence to perform molecular biological analysis qualitatively using modern tools but also expands the boundaries for further studies of the Pel1 enzyme. In particular, future directions of the research project may focus on the study of the mechanical properties of the protein, the analysis of its toxicity, and the kinetics of its interaction with pectin.
Reference List
Kermani, A.A. (2021) ‘A guide to membrane protein X‐ray crystallography,’ The FEBS Journal, 288(20), pp. 5788-5804.
Quantifying protein using absorbance at 280 nm (n.d.) Web.
UniProt (2021) UniProtKB — P27760 (PLY1_AMBAR). Web.
Wu, P., Yang, S., Zhan, Z. and Zhang, G. (2020) ‘Origins and features of pectate lyases and their applications in industry,’ Applied Microbiology and Biotechnology, pp. 1-14.