Introduction
One of the significant public health threats is unknown pathogens that can lead to the development of infectious diseases. The danger of such pathogens lies in the lack of knowledge about their nature and toxic properties and, thus, an understanding of the methods that can be used to control them preventively (Shiny Matilda et al., 2020). In other words, there is a need for qualitative identification techniques that aim to identify the bacterial pathogen strain. In microbiology, there are several identification techniques, among them staining procedures and metabolic tests.
The present laboratory work is structured around the examination of unknown bacterial specimens, the purpose of which is their qualitative identification. The laboratory experiment consists of an initial determination of the composition of the cell wall of the samples using the Gram staining method and then, based on the results, carrying out the appropriate tests. Thus, the present work describes the identification capabilities of microbiology for unknown bacterial specimens.
Materials and Methods
Two specimens (A and B), with identification number 44, as shown in Figure 1, were given by the professor before the tests were started. The nature of these bacterial specimens was not known and was to be ascertained initially through the use of Gram staining. Samples on blood agar were stained with crystal violet and, etched with iodine and then decolorized with ethanol; between each step, treatment with distilled water was performed to remove excess pigments.
Finally, safranin was used for re-staining: the Gram-positive bacteria retained their purple coloring, and the Gram-negative bacteria acquired the red safranin coloring. Microscopic examination was then performed for both types of bacteria, the purpose of which was to obtain information about the shape and arrangement of the bacteria in the specimen. A Catalase test was performed for Gram-positive coccus, a VP test was performed for Gram-positive rods, and a series of consecutive API 20 E tests were performed for Gram-negative rods.
The catalase test to determine the ability of the bacterium to synthesize catalase was performed by adding 3% hydrogen peroxide to a small amount of the sample. A positive reaction to the test corresponded to the release of gas bubbles because H2O2 was decomposed by catalase. Otherwise, no reaction was observed.
In addition, the SXT disk test, Camp text, Bacitracin and Optochin tests, Bile Esculin test, and NaCl test were performed for Gram-positive bacteria to identify the specific bacterial species. The SXT disk test was performed by placing a paper disk soaked with sulfamethoxazole-trimethoprim on the sample plate, and if positive, a clear zone of antibiotic resistance was observed around the disk. Camp text was performed by stroking the sample near Streptococcus agalactiae to determine the potential for hemolysis, and a positive reaction would be a tip-shaped zone of hemolysis.
Bacitracin and Optochin tests were performed by placing paper disks impregnated with each type of antibiotic on the sample plate to determine resistance to each type of antibiotic. The Bile Esculin test was performed by inoculating the bacterium in bile-esculin agar, and a positive reaction corresponding to the ability to hydrolyze esculin would correspond to the formation of a black pigment. Finally, in the NaCl test, the bacterium was inoculated on a salt medium (7.5% NaCl), and a positive reaction would be consistent with the ability of the bacterium to grow.
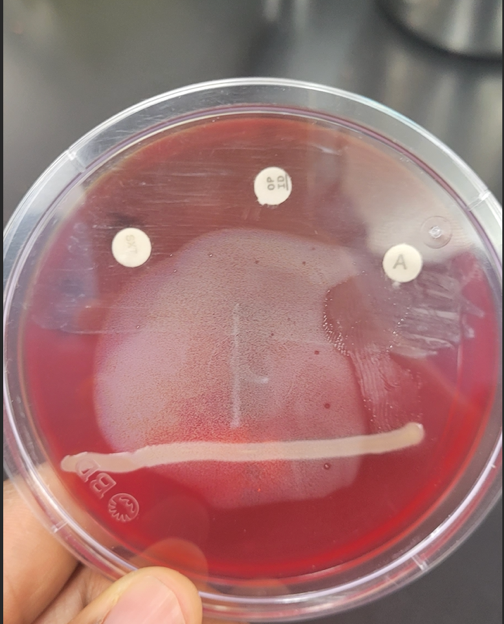
Results
During Gram staining and microscopic examination of the two samples, it was determined that Sample A belonged to Gram-negative rods without a pattern of bacteria in the colony, while Sample B was a Gram-positive coccus distributed as ordered chains (Table 1). Further laboratory work was performed only with Sample B since Sample A was given to a project partner.
Sequential microbiological tests were performed for Sample B, the purpose of which was to determine the ability to produce specific enzymes depending on the metabolic characteristics of the bacterium and the response to a specific test. Table 2 summarizes the data on each of the tests used. Two of the tests showed significant results, further demonstrated in Figure 2. In particular, Sample B gave a black positive reaction on the Bile Esculin Test and no bubbles, that is, no enzymatic process in the catalase reaction.
Table 1: Results of microscopic observation and Gram staining.
Table 2: Microbiological test results for Sample B.

Discussion and Conclusions
The purpose of the present experiment was to perform a qualitative identification of an unknown bacterium by performing microbiological staining techniques or metabolic tests. Gram staining showed that Specimen B was a Gram-positive bacterium and morphologically was a chain of located coccus. A series of tests were used for further identification.
For example, a positive reaction to the Bile Esculin Test confirmed the ability of the Specimen to hydrolyze esculin, which narrows the focus to group D streptococci and Enterococcus species (Aryal, 2022b). The catalase test showed a lack of catalase enzyme, which narrows the focus to Enterococcus or Streptococcus (Aryal, 2022a). In addition, a positive salt tolerance test indicated that Enterococcus (Aryal, 2022c) might be a candidate for the specimen. From all of the above, it follows that Specimen B belongs to the genus Enterococcus and is probably E. faecalis, E. zymogenes, E. liquifaciens, or E. durans.
A large number of different methods can be used to obtain more reliable results, but there is always a resource limitation in the real world. For clinical settings, such qualitative identification is of great benefit because it allows the nature of the potential pathogen to be identified and responded to more precisely and efficiently (Shiny Matilda et al., 2020). For example, bacteria of the genus Enterococcus are known to cause a large number of infections of varying severity, including urinary tract infections, heart valve infections, open wound and bloodstream infections, and meningitis (García-Solache & Rice, 2019).
For example, E. faecium causes wound infections, bloodstream infections, and urinary tract infections, including those transmitted in nosocomial settings. The multiplicity of pathological conditions caused by Enterococcus is realized by a large number of transmission routes: this includes direct contact and contamination through surfaces or food. Consequently, preventive measures are based on hygiene and sanitation practices, including hand and food washing and the use of a personal respirator when visiting public hospitals (García-Solache & Rice, 2019). If the patient has already been diagnosed with an infection caused by Enterococcus, antibiotics should be treated. Daptomycin can be used as an antibiotic treatment, allowing destruction of the bacterial cell membrane, or linezolid as an inhibitor of protein biosynthesis, in case of antibiotic resistance.
Although the experiment can be considered a success because all goals were met, the work was associated with some errors. All results have been tabulated, but in future works, it is recommended to make a photo registration in case of erroneous data and as an additional check. In addition, it is acceptable to refer to standardized methodological decision trees, which aim to use tests and staining consistently to identify a specific unknown specimen (Papagiannopoulou et al., 2020). The use of such strategies is expected to reduce the number of resources used and thus be more sustainable in terms of reagent consumption.
References
Aryal, S. (2022a). Catalase test — principle, uses, procedure, result interpretation with precautions. Microbiology Info. Web.
Aryal, S. (2022b). Bile esculin test — principle, procedure, uses and interpretation. Microbiology Info. Web.
Aryal, S. (2022c). Salt tolerance test – principle, procedure, uses and interpretation. Microbiology Info. Web.
García-Solache, M., & Rice, L. B. (2019). The Enterococcus: A model of adaptability to its environment. Clinical Microbiology Reviews, 32(2), 1-18. Web.
Papagiannopoulou, C., Parchen, R., Rubbens, P., & Waegeman, W. (2020). Fast pathogen identification using single-cell matrix-assisted laser desorption/ionization-aerosol time-of-flight mass spectrometry data and deep learning methods. Analytical Chemistry, 92(11), 7523-7531. Web.
Shiny Matilda, C., Madhusudan, I., Gaurav Isola, R., & Shanthi, C. (2020). Potential of proteomics to probe microbes. Journal of Basic Microbiology, 60(6), 471-483. Web.