Embedding genes into plasmid vectors is an integral part of molecular cloning as part of genetic engineering. This technique is widely used to obtain comparably cheap biological materials in a short time on a large scale, so genetic cloning and cell engineering techniques are spreading rapidly. One such example is the cloning of the pectate lyase gene using a plasmid vector, then inserted into prokaryotic organisms for culturing.
First and foremost, it should be said that pectate lyase is a naturally occurring extracellular enzyme responsible for the cleavage of pectin. Plant pectins, in turn, are polysaccharides formed by fragments of galacturonic acid, which help the plant retain the structure and maintain turgor pressure. The enzyme pectate lyase efficiently cleaves pectin by the beta elimination mechanism, which consequently inhibits plant stability. From the classification point of view, this enzyme belongs to the class of lyases that break covalent C-O bonds in polysaccharide molecules, which is accompanied by energy release. This functionality of the enzyme determines its biological significance: pathogenic bacteria and symbiotic bacteria of ruminants produce pectate lyase to degrade plant tissue and obtain nutrients. Laboratory cloning of pectate lyase using recombinant circuits makes it possible to create an enzyme that effectively catabolizes plant tissue degradation, including for faster maturation of soft fruits and textiles: hence, the economic value of this action is justified.
The molecular cloning procedure for this compound begins by isolating a gene from the original prokaryote that freely produces pectate lyase. For example, pectate lyase of the PL9 family produces the gram-positive Paenibacillus polymyxa KF-1 (Yuan et al., 2019). Another type of pectate lyase (PL1) is synthesized by cells of the ascomycete fungi Clonostachys rosea and Trichoderma (Atanasova et al., 2018). In either case, when a prokaryote or fungus suitable in economic, geographic, and pathogenic characteristics is selected, the gene responsible for the synthesis of this protein must be identified. This step is usually implemented using gene libraries, as sequences, locations, and even lists of suitable primers are published in databases, such as NCBI Genes. Thus, it is known that the protein sought is encoded by the pel1, pelA, PMR6, pelX, or a dozen other genes, depending on the organism from which the enzyme was obtained.
The pure gene must then be isolated from the organism using specially selected restrictions, which remove unnecessary fragments of the genome and retain only the gene of interest. Interestingly, prokaryotic cells, into which plasmids are then embedded rarely have their own intron removal mechanisms, and so biotechnologists often use the technique of complementary DNA. In this case, from a specific fragment of the eukaryotic RNA matrix that is able to synthesize pectate lyase, cDNA is cloned using a reverse transcription mechanism. The isolated gene must be amplified by PCR, which quantifies its presence many times over; otherwise, gene deficiency can affect the quality of molecular cloning. Amplification is the result of the PCR method, where short gene fragments are doubled with each cycle of the procedure using primers and free nucleotides. Confirmation of amplification is done by gel electrophoresis: an example of this is shown in Figure 1. This helps to understand that the PCR results show that the desired gene — with a previously known weight — has indeed been amplified and is present in the sample.
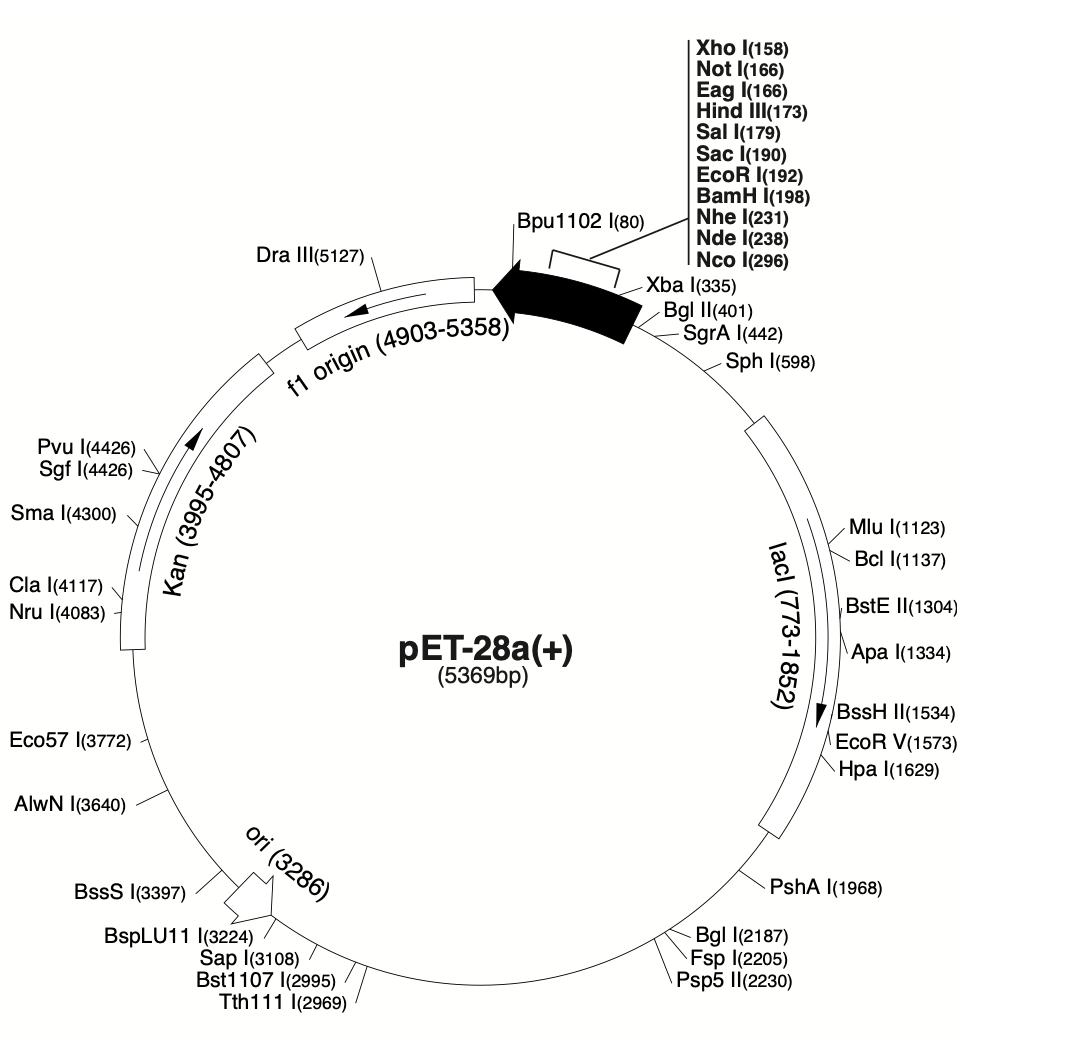
A sufficient amount of the collected gene must be transferred to a plasmid vector (pET-28a) in order to be then introduced into a prokaryote. Plasmids are independent circular DNA molecules that can be easily engineered. Plasmids typically carry genetic information about resistance to specific antibiotics. Such a molecule can be cut to insert new fragments or remove unwanted ones. Restrictions are used again here, but already to cut the plasmid; this allows the formation of sticky ends that will glue to the gene of interest isolated from the original organism. The pectate lyase gene has been shown to cut precisely by BamHI and XhoI (Zhao et al., 2018). DNA ligases are then used to “sew up” the unstable splicing and ensure the continuity of pET-28a.
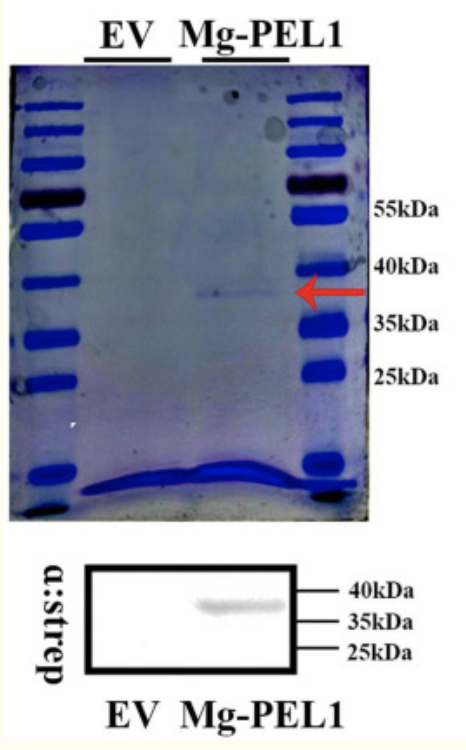
The generated expression vector can now be inserted into any organism that is easy to control and whose use is economically feasible, such as E. coli or yeast. The transfer of recombinant DNA is possible by means of a transformation process, whereby a strong thermal pressure provokes the bacteria to absorb foreign DNA. Since plasmids are responsible for antibiotic resistance in bacteria, the effectiveness of the incorporation of foreign plasmids into organisms in which these plasmids did not previously exist can be tested on colonies using the action of this antibiotic agent. Only those organisms that survived accepted the plasmids normally without destroying them and thus were able to synthesize pectate lyase.
References
Atanasova, L., Dubey, M., Grujić, M., Gudmundsson, M., Lorenz, C., Sandgren, M.,… & Karlsson, M. (2018). Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbiology, 18(1), 1-19. Web.
Chen, J., Li, Z., Lin, B., Liao, J., & Zhuo, K. (2021). A Meloidogyne graminicola pectate lyase is involved in virulence and activation of host defense responses. Frontiers in Plant Science, 12, 1-13. Web.
Novagen. (n.d.) pET-28a-c(+) vectors [PDF document]. Web.
Yuan, Y., Zhang, X. Y., Zhao, Y., Zhang, H., Zhou, Y. F., & Gao, J. (2019). A novel PL9 pectate lyase from Paenibacillus polymyxa KF-1: cloning, expression, and its application in pectin degradation. International Journal of Molecular Sciences, 20(12), 1-17. Web.
Zhao, Y., Yuan, Y., Zhang, X., Li, Y., Li, Q., Zhou, Y., & Gao, J. (2018). Screening of a novel
polysaccharide lyase family 10 pectate lyase from Paenibacillus polymyxa KF-1: cloning, expression and characterization. Molecules, 23(11), 1-17. Web.