Abstract
Microbial resistance to antimicrobial agents is a growing challenge for the development of chemotherapeutic agents. E. coli is a Gram-negative bacteria that contain pathogenic strains that lead to various illnesses including infections of the respiratory system, urinary tract and gastrointestinal system. Previous studies have reported that small molecules are being developed as chemotherapeutic agents in Staphylococcus aureus by targeting heme metabolism and iron. Since iron is also vital in the physiological reactions of E. coli and that changes in its concentrations affect the bacteria, it is hypothesized that altering the regulation of iron in E. coli by interfering with the structure of the Fur gene will affect the metabolism of E. coli hence lead to the death of bacteria. Such a move will enable the identification of drug targets in E. coli as well as the development of potential therapeutic agents that will help fight microbial resistance to antibiotics. This idea fits the definition of chemical biology because the experiment focuses on understanding the chemical processes of mutations on the functions of the Fur gene in controlling iron uptake and utilization.
Background
Escherichia coli is a species of Gram-negative bacteria found naturally in the environment, food, and gastrointestinal tract of animals and humans. Most strains of E. coli are harmless. However, certain strains are linked with the occurrence of infections of the respiratory tract such as pneumonia, gastrointestinal infections and urinary tract ailments.
E. coli has shown an extraordinary tendency for acquiring resistance to antimicrobial agents (Hasan et al. 689). Therefore, drug-resistant strains are continually becoming a danger to public health. Most microbial agents lead to the death of bacterial cells by interfering with normal physiological processes of bacteria hence lowering their pathogenicity. Some of the target areas include bacterial cell walls and enzymes. Therefore, studies that look at small molecules on bacterial cells and are valuable tools for understanding the fundamental biology and pathogenesis of these bactreia and may provide leads in the discovery and development of novel therapeutic agents. The advancement of virulence and drug resistance in community settings emphasize the need for developing new antimicrobial compounds and target pathways.
Iron is vital to most living organisms due to its importance in various metabolic processes. For instance, it is a component of all heme enzymes including cytochromes and hydroperoxidase. Iron is also necessary in as a cofactor for enzymes that facilitate reproduction, metabolism, and defense against reactive oxygen species. Nevertheless, iron is not found readily and bacterial cells have to use a number of iron uptake systems to obtain it. Despite the usefulness of iron, its concentration in bacterial cells needs to be kept within certain levels. Exceeding these levels leads to toxic effects within the cells, for example, the generation of reactive oxygen species that lead to the peroxidation of lipids within the cell membrane, damage to proteins and DNA (Andrews, Robinson and Rodriguez-Quinones 226). A study by Sorokina et al. (439) demonstrates that optimal growth was observed in E. coli at iron concentrations ranging from 0.1 to 2 mg per liter. Conversely, iron concentrations of 29 mg per liter lead to the complete inhibition of the growth of E. coli. Iron cannot be used directly as an antimicrobial agent because altering the concentrations in a human host will also have deleterious effects on the host. Previous studies have successfully demonstrated the use of heme homologs in disrupting heme metabolism in Staphylococcus aureus through interfering with heme sensor system (HssRS) that overcomes heme toxicity (Dutter et al. A).
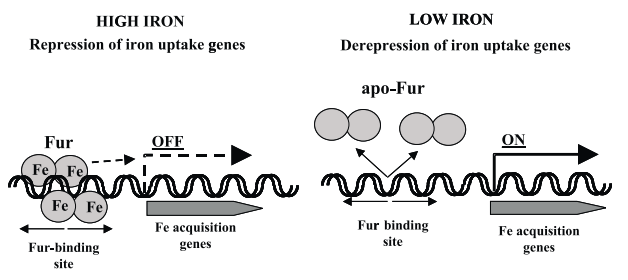
It is known that ferric-uptake regulator protein (Fur) controls the amounts of free iron in the intracellular space by regulating iron acquirement and iron usage, for instance, iron storage. Fur is a homodimer made up of 17-kDa subunits (Perard et al. B).
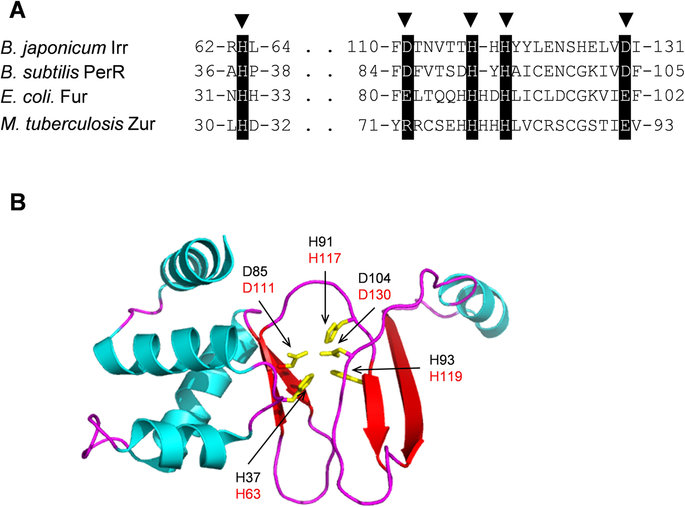
It functions as a positive repressor because it interacts with iron ions leading to the repression of transcription. Conversely, in the absence of iron ions, it brings about de-repression of transcription. It binds one ferrous ion for each subunit as well as other chemically-related ions such as cobalt and manganese. The attachment of a metal ion raises the affinity of the protein for its DNA-binding site by approximately 1000 times. In the absence of Fur, iron intake and utilization become imbalanced leading to excessive amounts of free iron.
Experimental Plan
Interfering with the Fur gene is the most suitable way of interfering with iron metabolism in E. coli.
Isolation of the Fur Gene
The Fur gene will be isolated from E. coli K-12 cells using synthetic primers that will be synthesized using known gene sequences of the Fur gene that will be obtained from the NCBI database.
Site-Directed Mutagenesis of the Fur Gene
Mutations will be introduced to the Fur gene at three different points which are upstream, downstream and the middle region of the gene using polymerase chain reaction. Different primers with mutations will be designed to incorporate the desired changes, which will be insertions, deletions and substitution. The three different mutations will then be applied to each region of the Fur gene. The mutant genes will be ligated into the lac Z operon of a plasmid.
Transfection of E. coli Cells and Selection of Transformed Cells
The plasmids will be incubated with E. coli cells. The transformed cells will be selected by growing the incubated cells in culture medium containing lactose due to the presence of the lac Z operon in the plasmids.
Confirming the Efficacy of the Mutation
The transformed E. coli cells will then be grown in medium containing various concentrations of iron to determine the activity of the mutant Fur genes in regulating iron uptake and usage in unaltered E. coli as the negative control.
Significance and Summary
Carrying out this project will provide knowledge regarding the effect of introducing mutations into the Fur gene on iron uptake regulation and its effect on the growth of E. coli cells. Other E. coli genes such as acnA, bfr, ftnA, fumA, fumB,
sdhCDAB, and sodB are also activated by iron in a Fur-reliant fashion. Carrying out site-directed mutagenesis at three different points will provide data that will facilitate the identification of mutation sites that lead to the complete inhibition of the Fur gene hence hampering iron metabolism. Mutants that will produce the largest inhibition effect will then be developed into therapeutic agents that will target iron metabolism in pathogenic E. coli. The effect of the Fur protein on the other genes will possibly produce a synergistic effect in preventing microbial growth.
Iron is an important element in the metabolism of bacteria and should be provided for a microbe to thrive and effect pathogenesis. However, the levels of iron concentrations should be maintained at given levels to prevent harmful effects of excessive iron. In pathogenic E. coli, interfering with the Fur gene, which is responsible for the regulation of uptake and utilization of iron, is a potential method of developing chemotherapeutic agents to combat microbial drug resistance.
Works Cited
Andrews, Simon C., Andrea K. Robinson, and Francisco Rodriguez-Quinones. “Bacterial Iron Homeostasis.” FEMS Microbiology Reviews. 27.2003 (2003): 215-237.
Dutter, Brendan F., Laura A. Mike, Paul R. Reid, Katherine M. Chong, Susan J. Ramos-Hunter, Eric P. Skaar, and Gary A. Sulikowski 2016, “Decoupling Activation of Heme Biosynthesis from Anaerobic Toxicity in a Molecule Active in Staphylococcus aureus.” ACS Chemical Biology.
Hasan, Badrul, Rayhan Faruque, Mirva Drobni, Jonas Waldenström, Abdus Sadique, Kabir Uddin Ahmed, Zahirul Islam, M. B. Hossain Parvez, Björn Olsen and Munirul Alam. “High Prevalence of Antibiotic Resistance in Pathogenic Escherichia coli from Large- and Small-Scale Poultry Farms in Bangladesh.” Avian Diseases. 55.4 (2011): 689-692.
Perard, Julien, Jacques Cove, Mathieu Castellan, Charles Solard, Myriam Savard, Roger Miras, Sandra Galop, Luca Signor, Serge Crouzy, Isabelle Michaud-Soret, and Eve de Rosny 2016, “Quaternary Structure of Fur Proteins, a New Subfamily of Tetrameric Proteins.” Biochemistry.
Sorokina, E. V., Yudina, T. P., Bubnov, I. A., and Danilov, V. S. “Assessment of Iron Toxicity Using a Luminescent Bacterial Test with an Escherichia coli Recombinant Strain.” Microbiology. 82.4(2013): 439-444.
Figure References
Andrews, Simon C., Andrea K. Robinson, and Francisco Rodriguez-Quinones. “Bacterial Iron Homeostasis.” FEMS Microbiology Reviews. 27.2003 (2003): 215-237.
Kitatsuji, Chihiro Kozue Izumi, Shusuke Nambu, Masaki Kurogochi, Takeshi Uchida, Shin-Ichiro Nishimura, Kazuhiro Iwai, Mark R. O’Brian, Masao Ikeda-Saito and Koichiro Ishimori. “Protein Oxidation Mediated by Heme-Induced Active Site Conversion Specific for Heme-Regulated Transcription Factor, Iron Response Regulator.” Scientific Reports 6.18703 (2016).
Wood, Marcia 2011. Lesser Known Escherichia coli Types Targeted in Food Safety Research.