Introduction
Physicochemical studies of key biological molecules are an essential part of laboratory practice to better understand their structure and identify possible patterns. The idea behind this work was to determine the viscosity of DNA molecules. It is well known that DNA is a long biopolymer consisting of four different nucleotides with different nitrogenous bases: Thymine, Adenine, Cytosine, and Guanine. In light of the enormous length of the DNA molecule localized in the nucleus of the body cells and the presence of double and triple hydrogen bonds that determine the spatial configuration of the polymer, it is reasonable to determine the factors responsible for the viscosity of bacterial DNA. In particular, a ring DNA molecule, also called a plasmid, of the prokaryote E. coli, was chosen as the study object. For the bacterium, the viscosity of the DNA was shown to be increased in solutions but minimal in intact cells. Moreover, the effects of thermal denaturation, precipitation, and mechanical shearing on this property were investigated. Consequently, the aim of this laboratory work was to conduct an in-depth empirical and comprehensive study of the nature of DNA viscosity and to identify the factors that determine it.
Methodology
The basic idea characterizing the experimental part of the laboratory work was to compare the release time of the cell suspension from the pipette between the control and experimental groups. For this purpose, a ten-milliliter Pasteur pipette was used with red and black markings on it, as shown in Figure 1. A sample of 9 mL of cell suspension was drawn into the pipette and, through a natural vertical release, was transferred to the test tube. The time it took for the sample to flow out was recorded in Table 1. After that, 1 mL of 10% SDS and 0.2 mL of 10M NaOH were added to the cell suspension to initiate the cell lysis process. Subsequent processes included mechanical shear induction, thermal denaturation, and precipitation used to assess the degree of effect on the viscosity of the DNA molecule.
Mechanical Shearing
This principle was based on successively passing the same sample of cell suspension through the Pasteur Pipette with the lysis mixture to evaluate the time of each pass. More specifically, 4 mL of suspension were loaded into the Pasteur pipette, and chemicals were added. The time required for the complete release of the suspension into a clean tube was recorded in the corresponding row of the Table, called the first shearing. The second shearing characterized the time it took for the products of the first pass to pass through the pipette again with the lysis mixture. The procedure was repeated until the time taken for complete release equaled that of the original base suspension.
Thermal Treatment
Preheating of cellular material was used to determine the effect of temperature on DNA viscosity. Specifically, 2 mL of cell lysate was sent to boiling water for ten minutes, after which it was cooled in an ice bath in a tube with a tightly screwed lid. The cooled cellular material was immersed in a Pasteur pipette to estimate the time of complete lysate passage. The result is recorded in the last row of the Table.
DNA Precipitation
A controlled interaction with ethyl alcohol was used to precipitate the DNA molecule chemically. Alcohol is known to cause the denaturation of molecules, and this postulate was used as an idea for viscosity testing. To 6 mL of cell lysate, 8 mL of ethyl alcohol was added. The expected differentiation of the layers occurred, and a glass rod was used for stirring. As a result, long white strands of denatured DNA were identified and coiled onto the test stick.
Results
Table 1. Data summarized the time required for each sample to be released entirely from the Pasteur pipette
The primary variable of this laboratory work was the time it took for the cell lysate suspension to pass through the Pasteur pipette completely. Consequently, the entire array of results is presented in Table 1, in which each sample and method of processing corresponds to its own time. Primarily, it can be seen that the most significant time resources were required for the cell lysate samples, while the lowest number of seconds for complete passage was for the boiled cells and the sample that underwent the fifth phase of mechanical shearing. For a better visual representation of the results, it was appropriate to interpret them as a diagram shown in Figure 2.
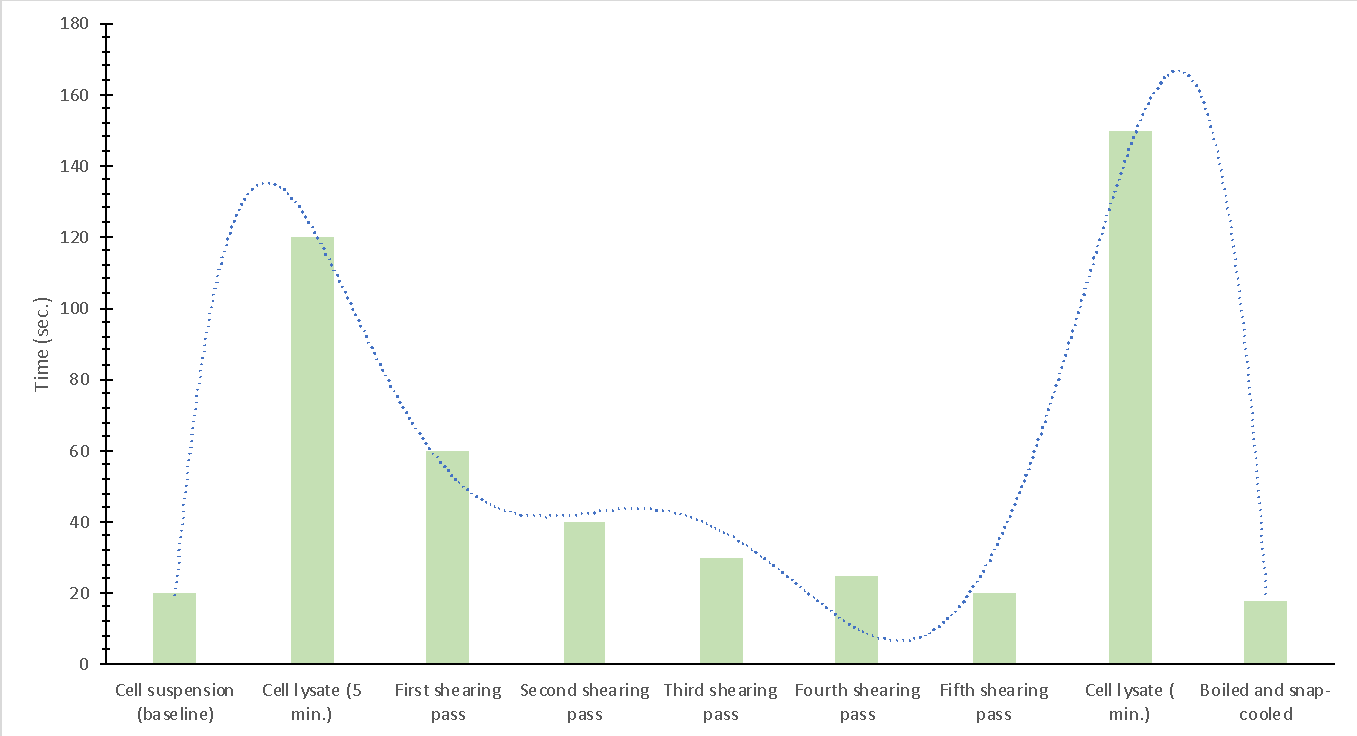
Figure 2 gives an idea of how the cell suspension processing techniques used are related to the material’s viscosity. Thus, the greater the DNA molecule’s viscosity, the longer it took for it to pass through entirely. This was evident in the first two lines of the experiment, when the solution’s viscosity was increased during the chemical lysis of the cells, with the time to pass increasing by a factor of 6 relative to the baseline line. In a series of mechanical shearing tests, it was shown that when the order of the procedure was increased, the DNA’s viscosity naturally decreased, indicating that the lysis had passed. Thus, lysis of the cells was shown to influence the decrease in the viscosity of the mixture positively. This assumption is also confirmed by the analysis of thermal denaturation when heating caused the double helixes of DNA molecules to break into chains, and the denaturing agents had low viscosity values. It is fair to note that this value was even lower than the baseline, which means that temperature strongly affects viscosity. In addition, the long white strands that appeared during the laboratory experiment were indicative of the deserialization of the cell chromosomes initiated by heating or the addition of toxic alcohol.
Conclusion
To summarize this work, DNA viscosity is a variable that can be manipulated. In particular, this test’s procedures, namely mechanical shearing, thermal denaturation, and deposition, showed that DNA viscosity decreases when hydrogen bonds are broken. This was verified by measuring the time of passage of the cellular material through the pipette. Thus, it can be postulated that the denaturation of molecules reduces the viscosity of DNA.