Aim
The aim of the present work was to determine the purity of the isolated cellular DNA sample from E. coli obtained in previous tests. The DNA is a double helix linked complementary by hydrogen bonds, with a phosphodiester bond implemented between the nucleotides of one strand. This bond could easily be broken by the addition of the enzyme DNase to the sample, causing the genetic molecule to separate into fragments. The purity of the sample in the laboratory was determined by analyzing such fragments. In particular, placing a DNase-treated and an untreated enzyme-treated sample on the gel electrophoresis matrix allowed the identification of differences in their migration when an electric current was applied since DNase completely destroyed the fragments, making it impossible to detect them in the corresponding track of the plate. This method was excellent for determining the purity of cellular material, from which the hypothesis of the entire laboratory experiment was that treatment of the sample with DNase would indeed result in the complete destruction of any traces on the gel-agarose plate.
Method
The methodological basis for the experiment was to use a cell sample previously isolated from E. coli to determine its purity. Based on the data from the table prepared in the ninth laboratory work, DNA extract, buffer solution, distilled water, and DNase were added to a 1.5-mL microcentrifuge tube. The tube was placed in a microcentrifuge to stir the contents vigorously, allowing larger particles, including impurities, to settle to the bottom. The contents were incubated for 10 minutes at 37°C, after which Stop/Loading Dye containing EDTA, glycerol, and Bromophenol Blue dye was added to the tube. The addition of this dye was necessary, in particular, to provide color identification on gel electrophoresis because the DNA molecules themselves are colorless and, therefore, invisible to the naked eye. After the addition of the dye in the ultraviolet irradiation, the molecules acquired a characteristic orange color, which greatly simplified the identification procedure. Based on the results obtained, the picture of gel electrophoresis, conclusions were to be drawn about the effect of DNase on the cell sample from E. coli when compared with the negative control, in which the enzyme was not added.
Result
The main result of the present laboratory work is a picture of gel electrophoresis, which allows the identification of the samples depending on the methods of treatment. This picture is shown in Figure 1, and it should have been clear for qualitative and quantitative separation of the samples from each other. However, referring to Figure 1 demonstrates the vagueness of the gel electrophoresis pattern, and thus no separation is possible. Both wells in which the treated and untreated cell samples were loaded showed stains, and these stains were unformed.
Discussion
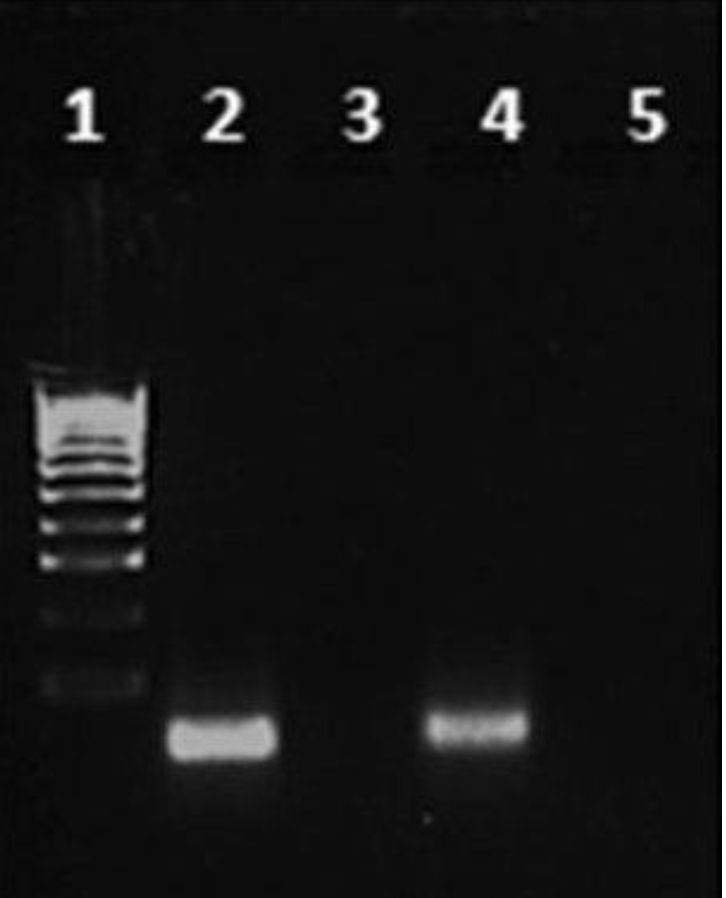
The purpose of this work was to qualitatively identify a cellular sample of DNA isolated from E. coli using gel electrophoresis technology. At the end of the work, it should have been easy to determine the differences between the samples from the gel electrophoresis pattern because, as the study material suggests, the DNase enzyme treatment of the DNA should have resulted in the absence of any staining on the matrix, as shown in Figure 2. This would be because the DNase enzyme hydrolytically destroys the phosphodiester bonds in the DNA, which leads to the breakdown of the molecule. Thus, DNA destruction would result in the absence of the observed features on the gel electrophoresis matrix. At the same time, for the DNA fragments loaded into the wells, when an electric current was applied, differences in the migration of the molecules to the anode would have been observed. This differentiation would occur because molecules with a lower molecular weight would migrate to the anode faster than heavier molecules.
In reality, however, no visible signs of a flowing gel electrophoresis reaction were observed, as shown in Figure 1. Cellular samples of DNA treated and untreated with the enzyme did not migrate across the matrix and did not differ from each other. There are several reasons why the result may have failed to work for the data. First, the loading of the cellular material after microcentrifugation may have failed because probably more conductivity buffer solution needed to be added. As one reason for the failure of the experiment, the application of a high current could also have distorted the results. In addition, photographs of gel electrophoresis from another group of students were used for the work because most of the groups attempted gel electrophoresis identification was unsuccessful. To remedy this situation in future trials, it is recommended that more buffer be used when loading materials into wells and that copies of the gel electrophoresis matrix be used to ensure reliable results.
Reference
Ahmad, A. A., Workie, B., & Mohamed, A. A. (2020). Diazonium gold salts as novel surface modifiers: what have we learned so far? Surfaces, 3(2), 182-196.