Materials and Methods
GST Pull Down
The protein-protein interaction experiment produced the 6*His-USP elution fraction that was carefully transferred into a microfuge labeled USP7D. The labeled microfuge was then stored on ice. GST and GST-EBNA 1 peptide columns, which were initially loaded in the freezer, were obtained and allowed to thaw at RT for approximately 2minutes after which they were placed on ice. The lids and plugs from the peptide-loaded columns were removed and placed on#3 glass test tubes. The columns were washed twice with 1ml of 1*PBS PH 7.4.The aim of the pull down experiment was to use chromatography to find out whether there was any interaction or any biological activity between the two proteins (Hayes, Flanagan & Jowsey, 2005).
To ensure there was a positive interaction between the proteins 6*His USP7 and GST-EBNA1, two consecutive experiments were conducted. First, they were incubated for ~90 minutes in a fridge. Half of the purified 6*His-USP 7 (200ml) was dialyzed and mixed with 800 µL of 1*PBS pH 7.4 During this step, the SDS-PAGE gel was casted which took ~2hours (Reed, Holmes, Weyers & Jones, 2007).
The liquid fractions were collected and put in microfuge tubes labeled GSTE PD-UP and GST PD-UP which were then stored in ice. The liquid contained the unbound protein. The samples were centrifuged at 1000rpm for 1minute RT. Samples 7 and 8 were made by separating the liquid into two (20-µL) aliquots, which were similarly labeled. The samples were washed for six times. The sixth wash was obtained and labeled as GST-PDW and GSTE-PDW that is sample 9 and 10 respectively.
200 µL of a GST-Elution buffer were added to the samples after which they were incubated for 10 minutes. The Elutes were then transferred into microfuge tubes labeled GSTPD Elt and GSTEPD Elt and placed on ice. These tubes held the eluted fractions from the pull down extraction. They were labeled and represented samples 11 and 12 for SDS page analysis. SDS-PAGE Gel was casted and incubated. These samples were labeled as GST-PDUP and GSTE PD-UP respectively.
Table 1
(Middelberg, 2006)
Table 2
(Middelberg, 2006)
Results
The relationship between two protein produces 6* His-USP elution which was first put in microfuge (labeled USP7D) for incubation. GST and GST-EBNA 1 columns were obtained and allowed to thaw for about 2 minutes. The columns were then buffered twice with 1 ml of 18PBS pH 7. 4 (Reed, Holmes, Weyers, & Jones, 2007).
In order to ensure positive results, two respective experiments were conducted. Both setups were incubated for 90 minutes in a fridge. Part of it was dialyzed and mixed with 800 micro-liters of 1*PBS pH 7.4. At this stage, SDS-PAGE gel was casted. This experiment took approximately 2 hrs (Hong, Yu, & Kang, 2002; Atkinson & Babbitt, 2009).
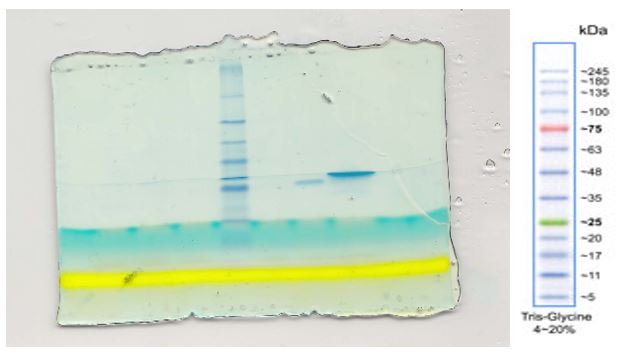
15% SDS PAGE Gel GST EBNA1 appears larger in the 6th lane of the legend figure the GST PD
Liquid portions were collected, labeled and placed in ice. They were then buffered for six times and re-labeled again as sample 9 and 10. 200 micro-liters of GST elution buffer were added to the samples after a 10 minutes incubation period. They were then labeled as sample 11 and 12 for SDS page analysis. A total of 12 samples were obtained from various experiment for the SDS-PAGE experiment. The samples were placed on different wells on the gels as shown in the tables above.
From legend figure, lane 8 constituents included
- GST pull down
- GST PD-UP
- GST-PDW
- Protein marker
- GSTE PDW
- GST-PD-Eit
- GSTE PD-Eit
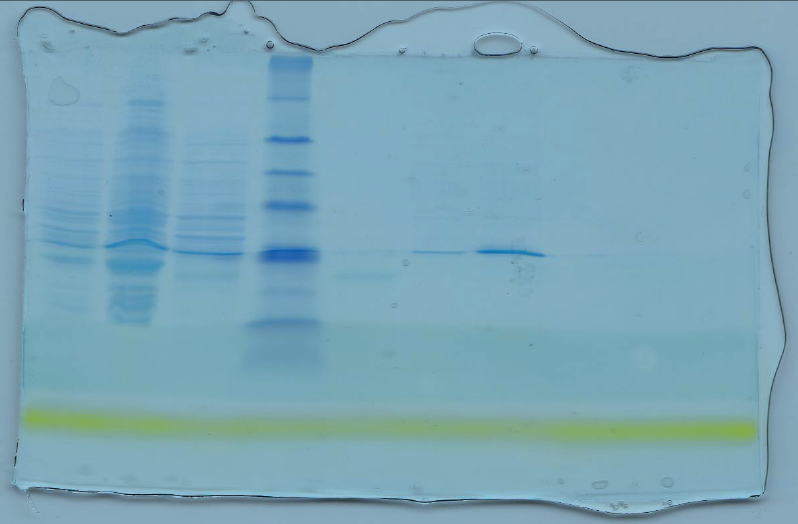
(Assay of protein pull-down, the interaction between 6* His-USP and GST-EBNA1 was studied. The procedure aimed at examining if there was any biological protein-protein interaction between the two proteins. A blue eye prestained protein was used as the control marker).
Discussions
From the above figure 2, the lane with GST EBNA produced a larger band compared to lane 6 that contained GST PD Elt. Virtually every cell in the human body contains an enzyme called Glutathione S-transferase (GSH). The enzyme is important especially for scavenging of free radicals through the below equation (Hong, Yu & Kang, 2002; Osuna, & Casamayor, 2011; MacDonald, & Lucy, 2006; Oakley, 2011).
2 GSH (reduced glutathione) ⇒ GSSG (OXIDISED GLUTATHIONE)
GSH is one of the GST’s substrate and hence GST can bind the unusual tri-peptide present in GSH with a very high affinity. The aim of the experiment was to exploit this high affinity interaction between the two compounds. The experiment involved initial purification of the two proteiens and later a set up to study their particular intterraction through a pulldown protein assay technique.The GSH can be coupled to sepharose or agarose beads, after which the beads can be employed in the affinity chromatography. Affinity chromatography can utilize either a gravity flow purification or a batch purification. This fusion however requires glutathione to be in its reduced GSH form. The GST tag is recovered by the application of a protease thrombin, an enzyme that that lyses cleaves at the thrombin cleavage site. The protein of interest is then obtained from GST (Atkinson & Babbitt, 2009; Wu, & Koiwa, 2012).
The ubiquitin protease, USP 7 is a deubiquitylating enzyme that removes ubiquitin from its substrate. It performs an important role in tumor suppression as it supports the function of the protein p53. EBV is on the hand a virus, which is greatly associated with some forms of cancer. EBV competes with p53 for USP53. The virus EBNA1 used in the experiment has been previously associated with effects that are similar to EBV. The study tested if the protein EBNA1 and USP interact (Rath, Glibowicka, Nadeau, Chen & Deber, 2009; Board, 2011).
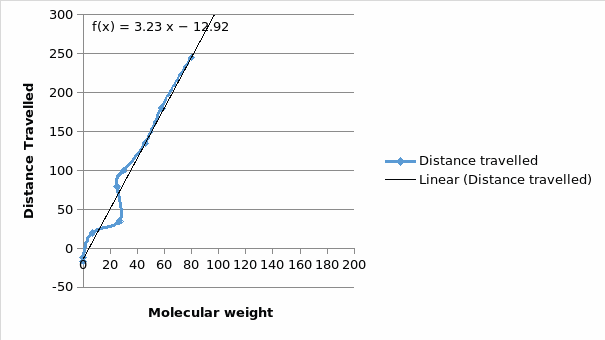
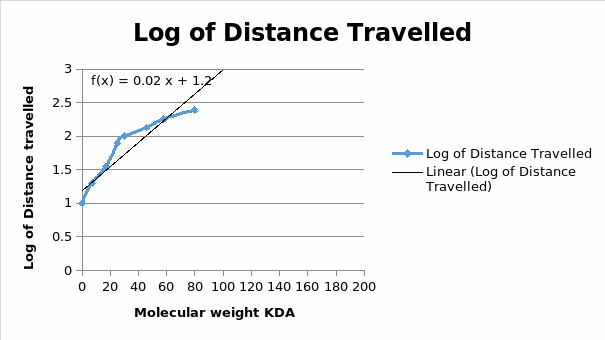
The blue bands on the clear background show that the procedure was perfect on figure 1. Figure 2 however has many errors, which might have been caused by contamination or by protein degradation. There is a direct relationship between a protein mobility and the log its molecular weight (Figure 5). The other drawback of such a purification is that if a protein requires post-translational modifications, the whole process definitely fails. The apparent molecular weights are most likely closer to the true protein weights. As was expected, there was reported a likely interaction between , USP 7 and the EBNA1 proteins. The standard curve requires the determination of the position of each band (Middelberg, 2006).
In conclusion, It is vey important that due to the many different proteins, any form of error be avoided during the protein sepataion. Accuracy is very important as it eliminates some of the errors that may lead to wrong results. Secondly, contaminations have to be avoided by ensuring use of sterile equipment. It is advisable to take note of the false positives and false negatives that may result.
References
Atkinson, H., & Babbitt, P. (2009). Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry, 48(46), 11108–11116.
Board, P. (2011). Glutathione transferases. Drug metabolism reviews, 43(2), 91.
Flanagan, J., & Smythe, M. (2011). Sigma-class glutathione transferases. Drug metabolism reviews, 43(2), 194 – 214..
Hayes, J., Flanagan, J., & Jowsey, I. (2005). Glutathione transferases.” Annual review of pharmacology and toxicology, 45(1), 51 – 88.
Hong, S., Yu, J., & Kang, S. (2002). Ultrastructural localization of 28 kDa glutathione S-transferase in adult Clonorchis sinensis. The Korean Journal of Parasitology, 40(4), 173 – 176.
MacDonald, A., & Lucy, C. (2006). Highly efficient protein separations in capillary electrophoresis using a supported bilayer/diblock copolymer coating. Journal of Chromatography A, 1130(2), 265 – 271.
Middelberg, A. (2006). Biomolecular Engineering. Chemical Engineering Science, 61(3), 875.
Oakley, A. (2011). Glutathione transferases: a structural perspective. Drug metabolism reviews, 43(2), 138 – 151.
Osuna, B., & Casamayor, E. (2011). Sodium Dodecyl Sulfate-Polyacrylamide Gel Protein Electrophoresis of Freshwater Photosynthetic Sulfur Bacteria. Current Microbiology, 62(1), 111 – 116.
Rath, A., Glibowicka, M., Nadeau, V., Chen, G., &Deber, C. (2009). Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proceedings of the National Academy of Sciences, 106(6), 1760–1765.
Reed, R., Holmes, D., Weyers, J., & Jones, A. (2007). Practical Skills in Biomolecular Science (3rd ed). Toronto: Pearson Education Canada.
Wu, X., & Koiwa, H. (2012). One-step casting of Laemmli discontinued sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. Analytical biochemistry, 421(1), 347.