Therapeutic drug monitoring (TDM) refers to the measurement of chemical or biochemical parameters in the lab to inform drug prescribing procedures. The precise parameters that are measured in TDM include drugs and their metabolites (Baktir 2017). These measurements are usually taken at predetermined intervals following the administration of a drug to sustain a constant concentration in plasma. The therapeutic range of a drug denotes the extent of dosage or plasma concentration that elicits the required treatment effect. It is defined by an upper and lower limit of the proportion between toxic and effective dosages of medication. The bigger the range between the two values, the larger the therapeutic index and the safer the drug and vice versa. TDM is critical for drugs with a narrow therapeutic index. Examples of such medications include cardiac drugs like digoxin and lidocaine, antibiotics such as aminoglycosides, chloramphenicol and vancomycin, bronchodilators such as theophylline, antiepileptics such as phenobarbital, carbamazepine, and phenytoin, anti-cancer medications such as methotrexate, and immunosuppressants such as tacrolimus and cyclosporine (Baktir 2017; Clarke 2020).
The pharmacokinetic and pharmacodynamic capabilities vary from one individual to another and determine the overall effectiveness of medications. Different people will absorb, metabolise, use and excrete medications at varying rates. Some of the variables that determine the rate and extent of drug metabolism include age, genetic makeup, body weight and general health status. Therefore, it may be necessary to customise the dosages of specific medications to achieve the desired therapeutic outcomes. TDM provides an avenue for the tailoring of drug regimens with narrow therapeutic indices to match patients’ physiological requirements, thereby avoiding drug toxicity. Additional applications of TDM include monitoring the efficiency of medications, patient compliance and studying drug to drug interactions. Consequently, several biological assays have been developed to facilitate TDM. The purpose of this paper is to describe various immunoassays used in TDM as well as their advantages and shortcomings.
Immunoassays in the Measurement of TDM Analytes
Immunoassays are qualitative biochemical techniques that use antibody-antigen reactions to determine the presence as well as the quantity of a macromolecule in a solution. The properties of antibodies that make them suitable for immunoassays include a large association constant (Ka) and highly specific binding. Consequently, antibodies can be used to detect and quantify analytes in complex biological fluids such as urine, blood, sweat, serum, saliva, or vitreous humor (Clarke 2020). Antibodies are a type of protein known as glycoproteins. They can bind with high specificity to synthetic or natural moieties known as antigens. There are five distinct classes of antibodies: IgG, IgM, IgA, IgD and IgE. However, the commonly used antibody in immunoassays is IgG. Figure 1 shows the general structure of an antibody. It is made up of two similar heavy chains joined to two identical light chains via disulfide bonds.
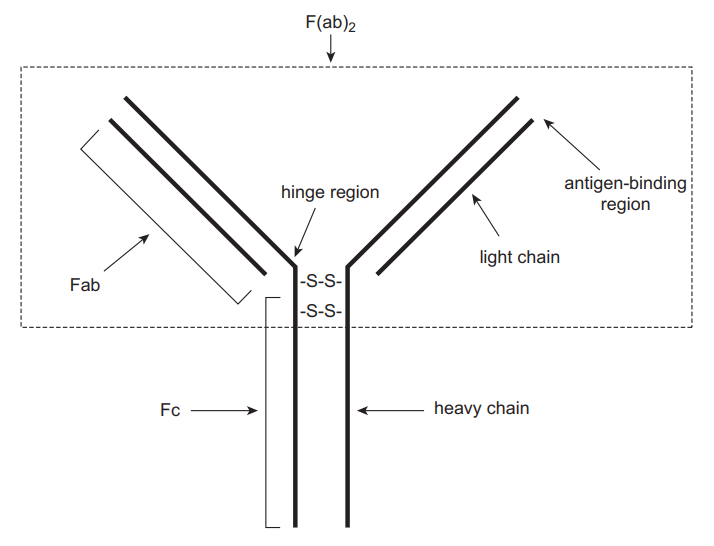
The Fc region does not have any antigen-binding capacity and is conserved in all antibodies. The (Fab)2 region consists of the antigen-binding zones and confers specificity to antibodies. The precise location on an antigen that an antibody binds is referred to as an epitope. Antibodies may be monoclonal or polyclonal depending on their origin. Polyclonal antigens are obtained from different cell lines. They target a particular antigen but at different epitopes. Therefore, their specificity may be affected, particularly if different antigens possess similar epitopes. Monoclonal antibodies, in contrast, are derived from a single cell line, which confers additional specificity because they target specific antigens at precise epitopes (Rosenstein et al. 2020). In immunoassays, chemical labels are often attached to antibodies or antigens to facilitate the detection of small quantities of an analyte. The types of labels or sequence of events followed in an assay determine the different forms of immunoassays. This section describes various immunoassays used in TDM.
Radioimmunoassays (RIA)
Radioimmunoassays use radioactive substances to label the analyte to be measured. Antibodies that are specific to the analyte are then added to the sample, which could be blood, serum, or any other biological fluid. Consequently, labelled and unlabelled analytes compete for the antigen-binding site of the antibodies. In the end, part of the labelled antigen does not bind to the antibody and is separated from the rest followed by detection by measuring its radioactivity. The amount of labelled antigen can be correlated to the quantity of unlabelled antigen, thereby finding its concentration as shown in Figure 2.
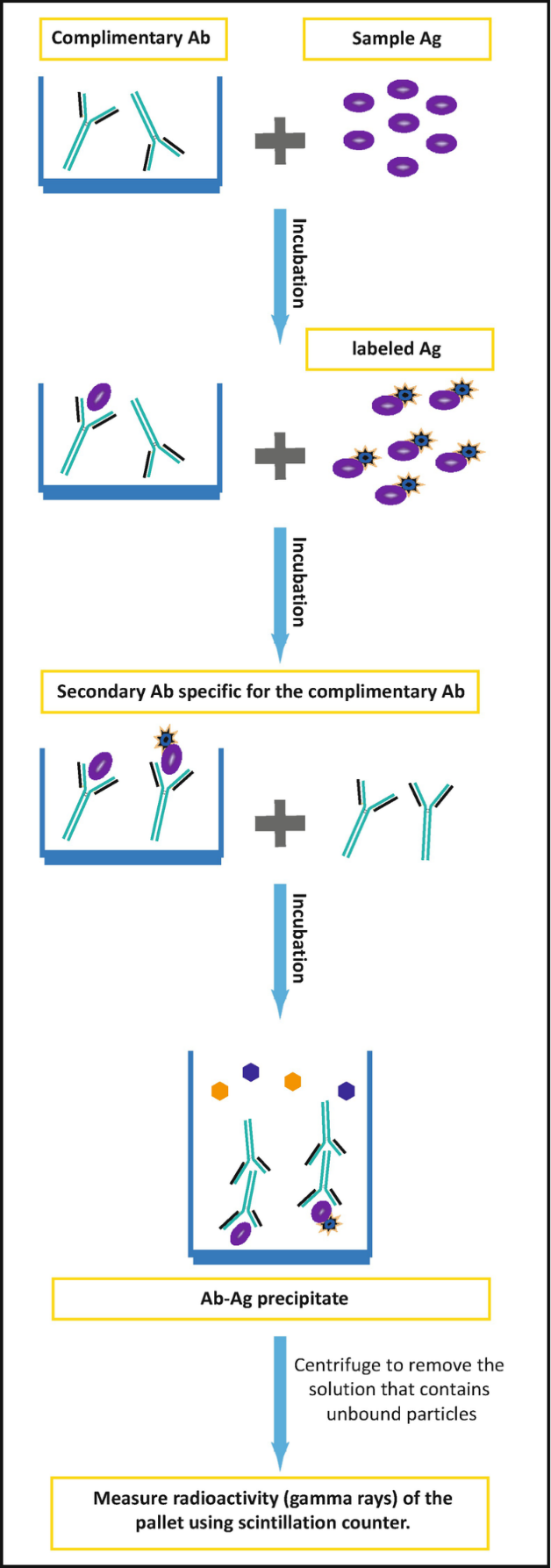
Some of the advantages of RIA include simplicity, high sensitivity and low background interference. However, its shortcomings include the requirement of a scintillation counter, which may not be readily available. Furthermore, exposure to radioactive substances poses health hazards (Alhabbab 2018). The process cannot be automated and takes long to complete a single assay due to the half-life of most radioactive agents. RIA is commonly used to monitor the levels of antibiotics such as tobramycin.
Fluorescence Polarisation Immunoassay (FPIA)
In fluorescence immunoassays, chemical agents that fluoresce (emit light at particular wavelengths) are commonly used to detect antigen-antibody complexes. Examples of fluorescent dyes used in fluorescence assays include fluorescein isothiocyanate (FITC), Lucifer yellow and Rhodamine B (Clarke 2020). The principle of competitive binding such as that used in RIA also applies to this technique. Antigens labelled with a fluorescent dye compete with the unlabelled antigen in the sample for antibody binding sites. Fluorescence polarisation immunoassay (FPIA) takes advantage of the polarisation of fluorescence as a result of the molecular revolution in solution to estimate the degree of antibody binding (Bouquié et al. 2016). Unbound, labelled antigens tend to revolve very fast, leading to a high degree of anisotropy and a low level of polarisation (Figure 3). The binding of a label lowers the extent of rotation and subsequent anisotropy, leading to a higher polarisation. The difference in polarisation is directly related to the amount of antigen in a sample.
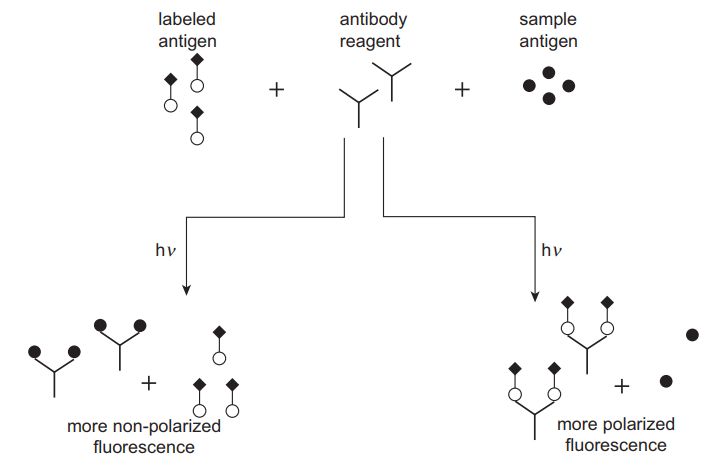
One advantage of FPIA is that no separation step is required because of the influence of antibody binding on the behaviour of antigens, making it faster. It is also sensitive in the detection of low analyte concentration and is safer than radioimmunoassays. However, the main disadvantage of the method is high background interference that is attributed to the inherent fluorescence of biological samples such as bilirubin, proteins, drugs themselves and their metabolites. This technique is used in the TDM of antibiotics such as gentamicin as well as immunosuppressants such as cyclosporine A.
Fluorescence Energy Transfer
This method applies the dual-labelling of antibodies such that an analogue is first attached to a donor fluorophore while an acceptor molecule is connected to the antibody. Physical proximity between the antigen and antibody results in the stifling of the fluorescence of the donor and energy conveyance to the acceptor. This way, antibody binding alters the signal of the label, thereby facilitating homogenous immunoassay (Clarke 2020). The advantages and shortcomings of this assay are similar to those in FPIA. This method has been used in the TDM of antiepileptics and cardiac drugs.
Enzyme-Linked Immunosorbent Assay (ELISA)
Enzyme-based assays use biological catalysts such as horseradish peroxidase, alkaline phosphatase, beta-galactosidase, or glucose-6-dehydrogenase. These enzymes are paired with substrates that react to produce a coloured product that can then be assayed. Often, the intensity of the coloured product can be correlated to the concentration of the analyte through photometric monitoring (Darwish, Al-Shehri, and El-Gendy 2018). A fluorescent product, for instance, umbelliferone may also be produced and assayed. In some cases, the initial enzyme-substrate reaction may produce an intermediate product that undergoes a second enzymatic reaction, leading to the amplification of the final signal and increased assay specificity. In ELISA, a sandwich immunoassay format is used where a capture antibody is immobilised to a solid support, for instance, antibody-coated wells, whereas the tracer antibody is coupled to an enzyme. A sample containing the antigen is added to the immobilised antibodies, leading to the formation of antibody-antigen complexes. A second enzyme-labelled antibody that is specific to the bound antigen (tracer antibody) is then added (Clarke 2020). Unbound antibodies are washed away before the addition of a substrate such as 3,3’,5,5’-tetramethylbenzidine. The quantity of the resultant product is related to the concentration of antigen in the sample. Modification of ELISA can be done to facilitate the detection of antibodies through the mobilisation of capture antigens instead of capture antibodies.
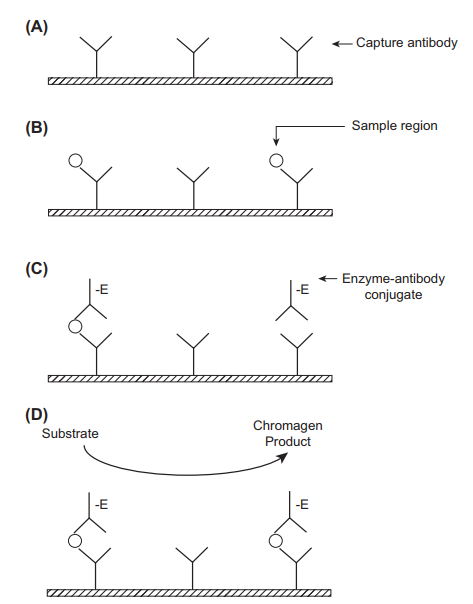
The advantages of ELISA include significant amplification of the signal by enzyme labels. A single enzyme label can amplify a signal 100 times, whereas a fluorescent substrate can result in a 100,000-fold signal amplification (Clark 2020). The technique is also fast, simple to perform, cost-effective and provides accurate results. There is potential for automation, leading to the analysis of multiple samples simultaneously. It is also possible to detect antigens as well as antibodies. The limitations of ELISA include an arduous assay procedure and poor sensitivity in the recognition of certain challenging biomolecules, for instance, microRNAs. The readouts must be taken within a limited time because they are based on enzyme-substrate reactions. ELISA is used in the therapeutic drug monitoring of cardiac drugs such as amlodipine.
Microparticle Enzyme Immunoassay (MEIA)
Microparticle enzyme immunoassay is another form of enzyme-based assays. Antibodies that are specific to the target analyte are attached to the surface of microparticles. The test sample and antibody-coated particles are incubated in the presence of enzyme-labelled antibodies. Following the incubation period, the particles containing the bound sample and label are centrifuged to separate them from unbound substances. The particles may also be bound to a glass fibre matrix. Nonspecific binding is eliminated by washing with a buffer, followed by the addition of a substrate. The ensuing signal is indirectly related to the concentration of drug or metabolite in the sample. MEIA is fast and easy to perform in routine monitoring of drugs. The main shortcoming of this assay is substantial cross-reactivity from drug metabolites, leading to the production of falsely elevated drug concentrations (Dasgupta 2016). MEIA is applied in the TDM of immunosuppressants such as sirolimus in transplant patients.
Particle-Enhanced Turbidimetric Inhibition Immunoassay (PETINIA)
This technique is also referred to as immunoturbidimetric assay. In this assay, microparticles are attached to the target analyte. Antibodies are added to the mixture in an ideal ratio, leading to the agglutination of microparticles and the formation of insoluble complexes. Consequently, the turbidity of the solution is increased, causing it to scatter light. Free drug particles from the patient sample compete for the antibody binding sites and reduce the extent of particle aggregation. The rate of inhibition of complex formation is related to the concentration of the analyte. The advantages of this technique include simplicity and obtaining accurate results within a short time. The outcomes of PETINIA assays are comparable to established ELISA assays (Usman & Hempel 2016). It is also flexible and can be automated to analyse multiple samples simultaneously. Another advantage of PETINIA is that proteins in the analyte attach to the Fc portion of the antibodies, causing optimal orientation for an agglutination reaction. The key shortcoming of PETINIA is an inadequate sensitivity and dynamic range to contend with other conventional methods like chemiluminescence. Drugs that can be analysed this way include antiepileptics such as carbamazepine.
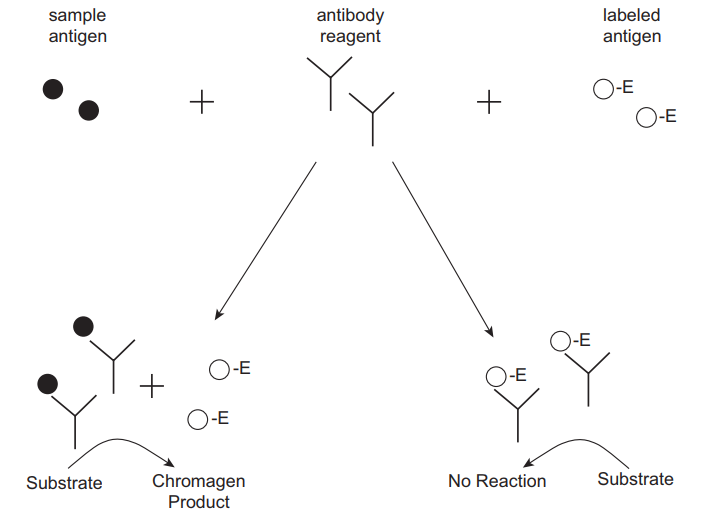
Enzyme-Multiplied Immunoassay Technique (EMIT)
A portion of the patient sample is incubated with an enzyme-labelled antigen in addition to a predetermined quantity of antibody that targets the antigen and a substrate. The labelled and unlabelled antigens (from the specimen) contend for antibody binding locations (Figure 5). The binding of the antibody to the labelled antigen inhibits enzyme activity because the interaction of enzyme and substrate is impeded. Alterations in the activity of the enzyme correlate to the amount of antigen in the test sample such that the concentration increases with a rise in enzyme activity (Shaikh & Guo 2017). The main strength of this assay is the capacity to quantify bound drugs without requiring separation from the unbound drug. A key shortcoming of EMIT is potential interference from cross-reacting constituents of the matrix. Medications that are usually monitored using this assay include antiepileptics.
Cloned Enzyme Donor Immunoassay (CEDIA)
Cloned enzyme donor immunoassay (CEDIA) is a competitive binding technique that exceeds EMIT assays in complexity. Two dormant fragments of the beta-galactosidase enzyme are produced through the genetic manipulation of the Z gene (lac operon) in E. coli. An active enzyme is generated by the reassembling of the two fragments even in the presence of covalent attachment to extraneous molecules (Clarke 2020).
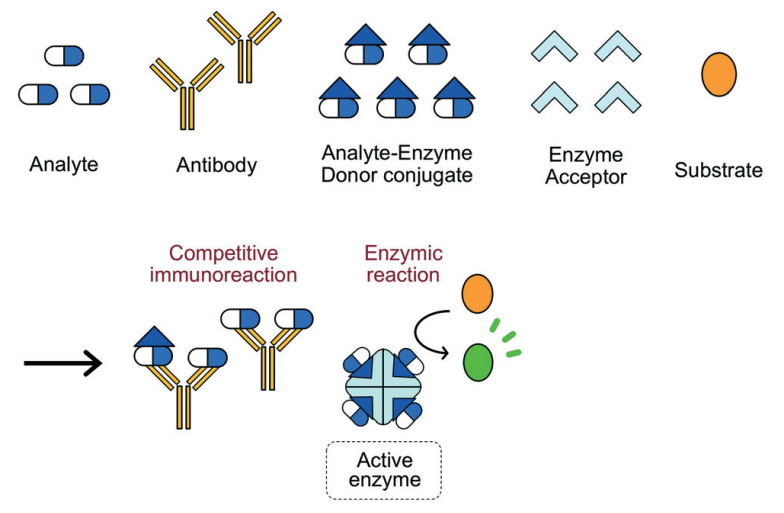
A competing antigen is labelled with one enzyme fragment and is incubated with the specimen, substrate, second fragment and an antibody against the target analyte. The binding of antibodies to labelled antigens precludes the reassembly of the complete enzyme hence no production of product from the substrate (Figure 6). The enzyme activity is proportional to the antigen concentration. Advantages include high sensitivity, accuracy and elimination of cross-reactivity (Sugiura et al. 2019). The main shortcoming of CEDIA is the possibility of false positives. This method is applied in the TDM of immunosuppressants, antiepileptics and bronchodilators.
Reference List
Alhabbab R.Y. (2018) Radioimmunoassay (RIA). in Basic Serological Testing. ed. by Alhabbab R.Y. New York: Springer, 77-81.
Baktir, G. (2017) ‘Therapeutic Drug Monitoring (TDM)’. Lectio Scientific 1 (1), 54-65.
Bouquié, R., Grégoire, M., Hernando, H., Azoulay, C., Dailly, E., Monteil-Ganière, C., Pineau, A., Deslandes, G., and Jolliet, P. (2016) ‘Evaluation of a Methotrexate Chemiluminescent Microparticle Immunoassay: Comparison to Fluorescence Polarization Immunoassay and Liquid Chromatography–Tandem Mass Spectrometry’. American Journal of Clinical Pathology 146 (1), 119-124.
Clarke, W. (2020) ‘Immunoassays for Therapeutic Drug Monitoring and Clinical Toxicology’. in Methods of Therapeutic Drug Monitoring including Pharmacogenetics. 2nd edn. ed. by Hampel, G. New York: Elsevier Science, 97-114.
Darwish, I.A., Al-Shehri, M.M., and El-Gendy, M.A. (2018) ‘Development of New ELISA with High Sensitivity and Selectivity for Bioanalysis of Bevacizumab: A Monoclonal Antibody Used for Cancer Immunotherapy’. Current Analytical Chemistry 14 (2), 174-181.
Dasgupta, A. (2016) ‘Limitations of Immunoassays Used for Therapeutic Drug Monitoring of Immunosuppressants’. in Personalized Immunosuppression in Transplantation: Role of Biomarker Monitoring and Therapeutic Drug Monitoring. 2nd edn. ed. by Oellerich, M. and Dasgupta, A. New York: Elsevier Science, 97-114.
Rosenstein, S., Vaisman‐Mentesh, A., Levy, L., Kigel, A., Dror, Y., and Wine, Y. (2020) ‘Production of F (ab′) 2 from Monoclonal and Polyclonal Antibodies’. Current Protocols in Molecular Biology 131 (1), 2-15.
Shaikh, A.S., and Guo, R. (2017) ‘Therapeutic Drug Monitoring of Phenytoin by Simple, Rapid, Accurate, Highly Sensitive and Novel Method and Its Clinical Applications’. Current Pharmaceutical Biotechnology 18 (13), 1098-1105.
Sugiura, K., Kaji, N., Tokeshi, M., and Baba, Y. (2019) ‘Development of a Microdevice For Facile Analysis of Theophylline in Whole Blood by a Cloned Enzyme Donor Immunoassay’. Lab on a Chip 19 (2), 233-240.
Usman, M., and Hempel, G. (2016) ‘Development and Validation of an HPLC Method for the Determination of Vancomycin in Human Plasma and Its Comparison with An Immunoassay (PETINIA)’. SpringerPlus, 5 (124), 1-7.